
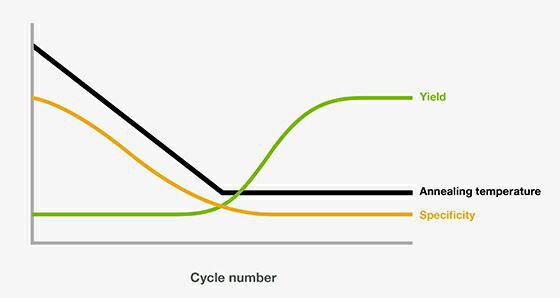
The best condition is determined empirically but there are situations where the variables are interdependent (such as varying the amount of dNTPs can affect the concentration of Mg 2+ in the reaction). However, there are merits in undertaking some minimal optimization steps that can improve yield and reproducibility, reduce non-specific amplification, as well as give a brighter product band with minimal background following gel electrophoresis.
While it is a good practice to optimize all the parameters in developing a new PCR protocol for the amplification of a new target, it might not be necessary or feasible to test all these variables each time. At this point, several parameters can be altered to enhance primer-template fidelity and product yield. Alternatively, amplification is successful but can lead to the generation of multiple, non-specific products, sometimes even excluding the desired product. Quite often, despite using an established protocol to amplify a specific region of DNA, the use of the same protocol for a different DNA region can result in failed amplification, even under optimal conditions. Despite being a powerful tool, it is not an exact science. Because of the great sensitivity of PCR, it has gained widespread popularity with a wide range of applications including gene cloning, DNA sequencing, mutagenesis, genotyping, assessing the expression level of a gene, and detection and diagnosis of pathogens. PCR is a powerful technique used to amplify a particular region of DNA so that it can be easily detected and manipulated. 3' terminus is extremely case sensitive - it must not be complementary to any region of the other primer used in the reaction and must provide correct base matching to template.Optimizing Strategies for PCR Amplification Success Inner self-complementary hairpins of >4 and of dimers >8 should be avoided. However it should be chosen empirically for individual conditions. Annealing temperature should be 5☌ lower than the calculated lower Tm. Calculated Tm for both primers used in the reaction should not differ >5☌ and Tm of the amplification product should not differ from primers by >10☌. Considerations in Primer Design GC-content should be between 40-60.The use of degenerate primers can greatly reduce the specificity of the PCR amplification.
#Designing touchdown pcr code
As several different codons can code for one amino acid it is not possible to design spefific primers. Degenerate primers are also used when the DNA sequence is unknown and primer design is based on the protein sequence. They may be convenient if the same gene is to be amplified from different organisms, as the genes themselves will be similar but not identical. These are mixtures of similar, but not identical, primers.


 0 kommentar(er)
0 kommentar(er)
